| Intro | History | Problems | Applet | Links | References |
|---|
HISTORY
For the sake of introduction, we'll simplify the model of study by reducing the dimensions to one, since it all started that way. And so instead of dealing with polygon, we will first look at permutations of sequence of numbers.
The motivation of the study of permutations arises in molecular biology and bioinformatics [3, 4, BP95, 5, 6]. One of the most dynamic field of research in science for the past decades has been the study of genomes and more precisely the human genome.
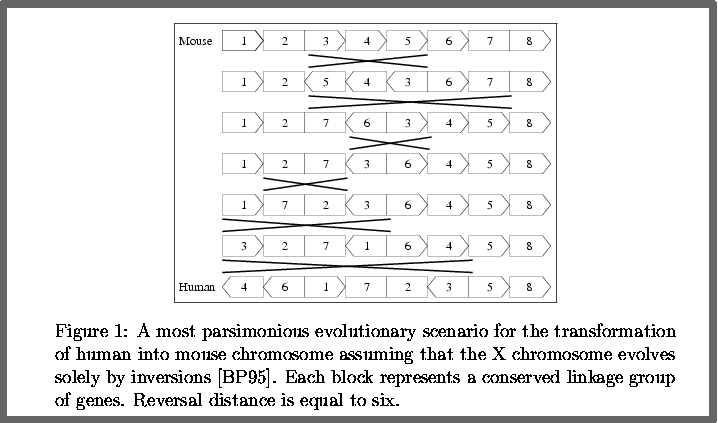
Concurrent with the fast progress of the Human Genome Projects, genetic and DNA data on many model organisms were accumulating rapidly, and consequently scientists were in urgent need of techniques for comparing genomes of different species.In the late Eighties, Jeffrey Palmer has demonstrated that different species have essentially the same genes, but the gene orders may differ between species. One of the best ways of checking similarities between genomes on a large scale is to compare the order of appearance of identical genes in the two species. Taking a distant perspective of genomes, the genes along a chromosome can be thought of as points along a line and can therefore be identified by signed numbers whose signs would correspond to their direction. Palmer and others have shown that the difference in order may be explained by a small number of reversals. These reversals correspond to evolutionary changes along the history between the species. Hence, given two such permutations, their reversal distance measures their evolutionary distance
Here's an example of how to compute the reversal distance between 2 sequences representing two gemones of different speacies.